Help
About YARG
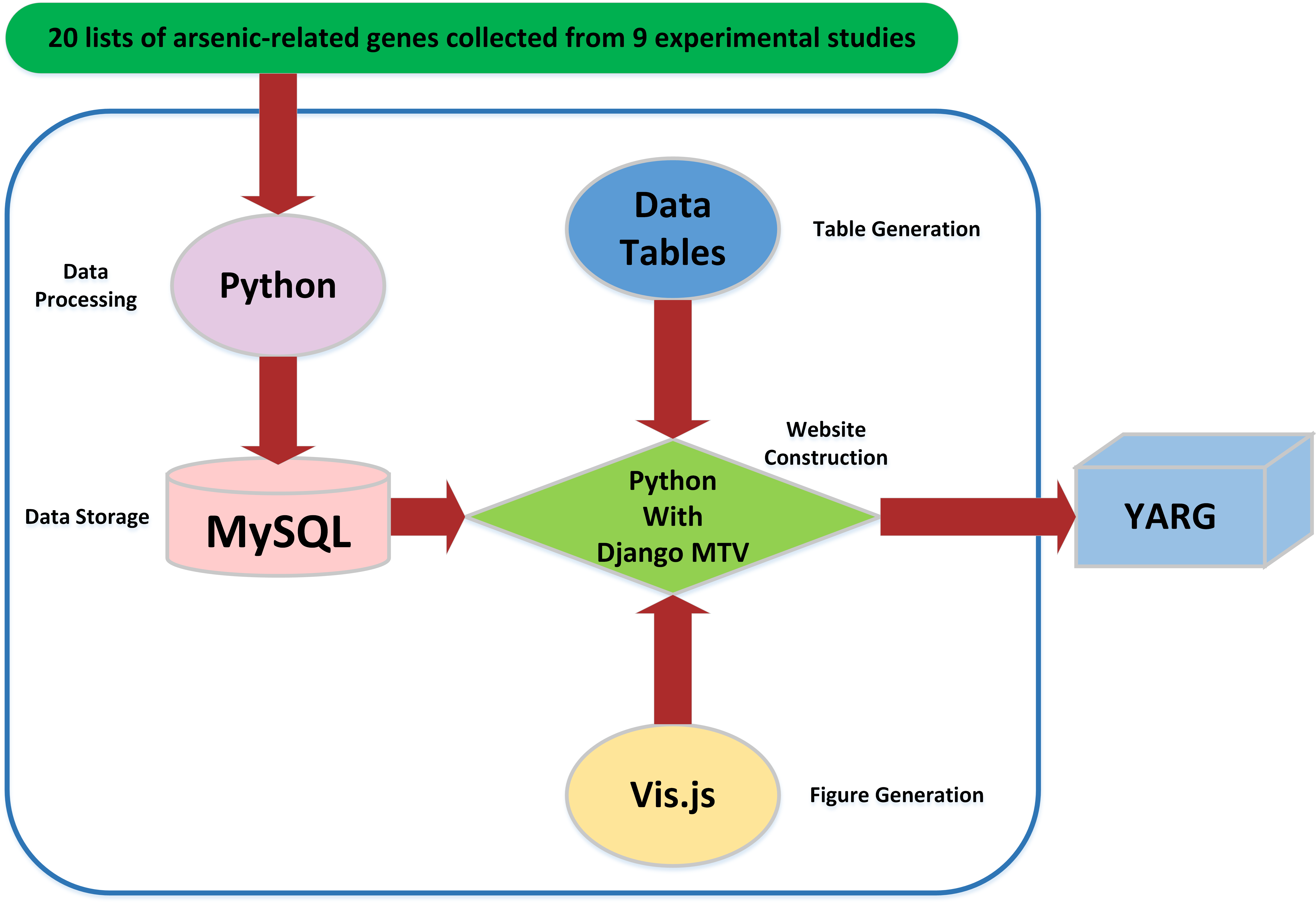
Construction of YARG
Usage of YARG
Motivation of YARG
Arsenic is a toxic metalloid. Moderate levels of arsenic exposure from drinking water can cause various human health problems such as blackfoot disease, circulatory disorders and cancer. Thus, arsenic toxicity is a key focus area for environmental and toxicological investigations. Many arsenic-related genes in yeast have been identified by experimental strategies such as phenotypic screening and transcriptional profiling. These identified arsenic-related genes are valuable information for studying arsenic toxicity. Unfortunately, they are scattered in many papers and researchers have no easy way to know this information. This prompts us to develop YARG (Yeast Arsenic-Related Genes) database.
What is YARG?
YARG (Yeast Arsenic-Related Genes) is a database which comprehensively collects 3396 arsenic-related genes in the literature. Users can search YARG by gene names to check whether they are arsenic-related genes and, if so, their arsenic-related experimental evidence from phenotypic screening or/and transcriptional profiling. Besides, users can browse YARG to retrieve 20 different lists of arsenic-related genes from nine experimental studies. The experimental strategy, experimental strain and experimental condition of these studies are also provided. Moreover, YARG has links to YeastMine to provide homology information. In summary, YARG is a useful resource for scientific community to investigate arsenic toxicity in yeasts and humans.
Configuration of YARG database
Python with the Django MTV framework was used to construct YARG website. Python was also used to do raw data processing. The processed data was stored in MySQL. The tables were produced by Data Tables (a table plug-in for jQuery). The graphics were generated by vis.js (a browser based graphic drawing library).
Collection of 3396 arsenic-related genes
We collected 20 gene lists in nine existing studies which experimentally identified arsenic-related genes by phenotypic screening (PS) or transcriptional profiling (TP). Among the 20 collected gene lists, 13 were generated by PS (see Table 1) and 7 were generated by TP (see Table 2). We then retrieved 3396 arsenic-related genes from these 20 collected gene lists.
Table 1. The 13 collected lists of arsenic-related genes identified by phenotypic screening
| Source | Identified gene list | Experimental strain | Experimental growth condition
(arsenic exposure) |
|---|---|---|---|
| Haugen et al. 2004 | 213 gene mutants of arsenite-sensitive phenotypes | Homozygous diploid S. cerevisiae BY4741 (MATa his3∆1 leu2∆0 met15∆0 ura3∆0) mutants | 100 μM and 1 mM sodium arsenite for 0.5, 2 or 4 h |
| Vujcic et al. 2007 | 72 gene mutants of arsenite-sensitive phenotypes | S. cerevisiae BY4741 (MATa his3∆1 leu2∆0 met15∆0 ura3∆0) mutants | YPD plates containing sodium arsenite (2.5 or 5 mmol/L) as well as on YPD plates without any arsenic (control plates). Plates were incubated for 4 to 15 days at 30°C and phenotype of each mutant was scored as sensitive or resistant compared with control plate and internal control (BY4741 on each plate). |
| Jin et al. 2008 | 65 gene mutants of arsenite-sensitive phenotypes | Homozygous diploid S. cerevisiae BY4743 (MATa/α his3∆1/his3∆1 leu2∆0 /leu2∆0 lys2∆0/LYS2 MET15/met15∆0 ura3∆0 /ura3∆0) mutants | 1.25 mM sodium arsenite for 2 h |
| Jo et al. 2009 | 647 gene mutants of arsenite-sensitive phenotypes | Homozygous diploid S. cerevisiae BY4743 (MATa/α his3∆1/his3∆1 leu2∆0 /leu2∆0 lys2∆0/LYS2 MET15/met15∆0 ura3∆0 /ura3∆0) mutants | 75, 150 and 300 µM sodium arsenite for 5 and 15 generations and subsequently analysed by TAG4 arrays. |
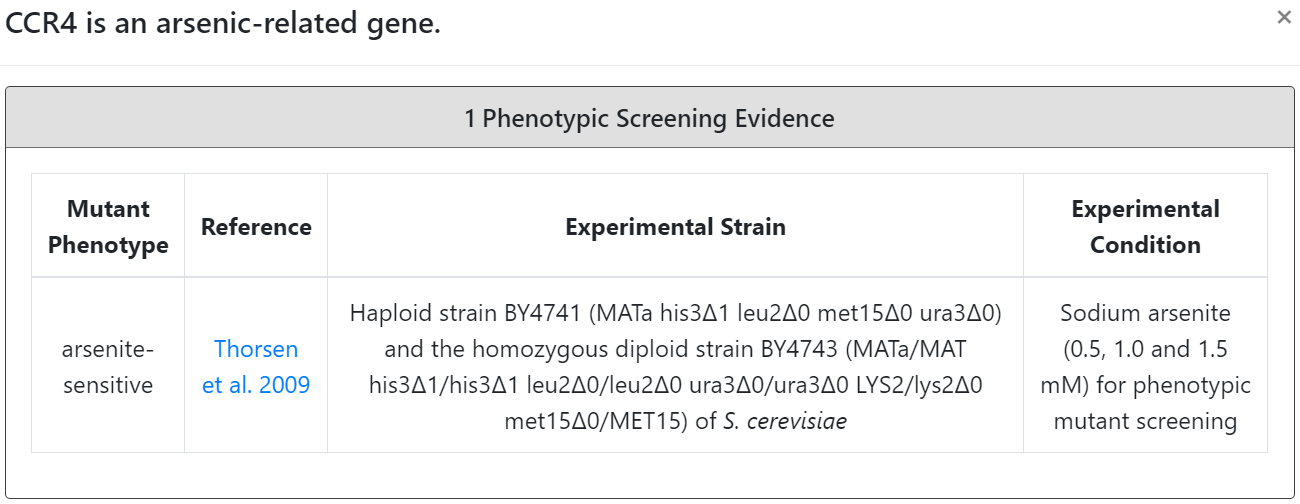
| Thorsen et al. 2009 | 305 gene mutants of arsenite-sensitive phenotypes | Haploid S. cerevisiae BY4741 and homozygous diploid S. cerevisiae BY4743 mutants | 0.5, 1.0 and 1.5 mM sodium arsenite for 24, 48 and 72 h |
| Zhou et al. 2009 | 245 gene mutants of arsenite-sensitive phenotypes | Haploid S. cerevisiae BY4741 mutants | 0, 0.75 and 1 mM sodium arsenite for 60 h |
| Pan et al. 2010 | 191 gene mutants of arsenite-sensitive phenotypes | Heterozygous diploid S. cerevisiae BY4741 mutants | 1 mM sodium arsenite for 1 h |
| Pan et al. 2010 | 33 gene mutants of arsenite-sensitive phenotypes | Heterozygous diploid S. cerevisiae BY4741 mutants | 450 mM sodium arsenite for 10 generations |
| Johnson et al. 2016 | 75 gene mutants of arsenite-sensitive phenotypes | Homozygous diploid S. cerevisiae BY4743 (MATa/α his3∆1/his3∆1 leu2∆0 /leu2∆0 lys2∆0/LYS2 MET15/met15∆0 ura3∆0 /ura3∆0) mutants | 0.2 and 0.4 mM sodium arsenite for 16 and 20 h |
| Zhou et al. 2009 | 5 gene mutants of arsenite-resistant phenotypes | Haploid S. cerevisiae BY4741 mutants | 0, 0.75 and 1 mM sodium arsenite for 60 h |
| Pan et al. 2010 | 109 gene mutants of arsenite-resistant phenotypes | Heterozygous diploid S. cerevisiae BY4741 mutants | 1 mM sodium arsenite for 1 h |
| Johnson et al. 2016 | 39 gene mutants of arsenite-resistant phenotypes | Homozygous diploid S. cerevisiae BY4743 (MATa/α his3∆1/his3∆1 leu2∆0 /leu2∆0 lys2∆0/LYS2 MET15/met15∆0 ura3∆0 /ura3∆0) mutants | 0.2 and 0.4 mM sodium arsenite for 16 and 20 h |
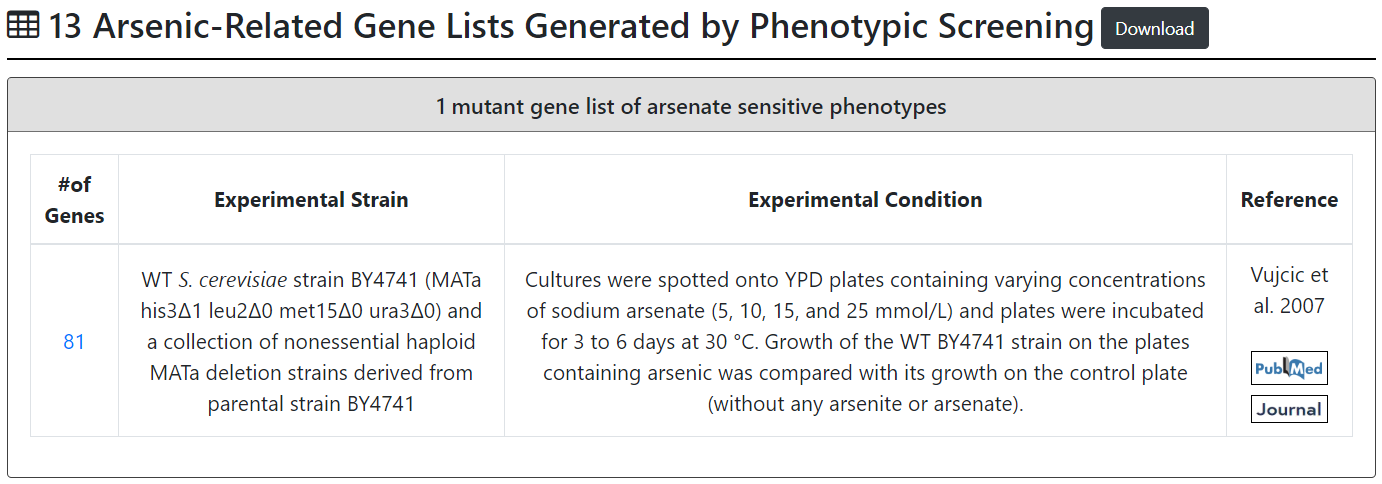
| Vujcic et al. 2007 | 81 gene mutants of arsenate-sensitive phenotypes | S. cerevisiae BY4741 and haploid MATa deletion mutant derived from parental strain BY4741 | YPD plates containing sodium arsenate (15 or 30 mmol/L) as well as on YPD plates without any arsenic (control plates). Plates were incubated for 4 to 15 days at 30°C and phenotype of each mutant was scored as sensitive or resistant compared with control plate and internal control (BY4741 on each plate). |
Table 2. The 7 collected lists of arsenic-related genes identified by transcriptional profiling
| Source | # of identified differentially expressed genes | Differentially expressed when comparing | Experimental strain | Experimental growth condition (arsenic exposure) |
|---|---|---|---|---|
| Thorsen et al. 2007 | 756 | WT 0.2 mM As(III) vs. WT | S. cerevisiae W303-1A | 0.2 mM sodium arsenite for 1 h |
| Thorsen et al. 2007 | 1066 | WT 1.0 mM As(III) vs. WT | S. cerevisiae W303-1A | 1.0 mM sodium arsenite for 1 h |
| Jin et al. 2008 | 1194 | WT 0.4 mM As(III) vs. WT | S. cerevisiae BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) | 0.4 mM sodium arsenite for 2 h |
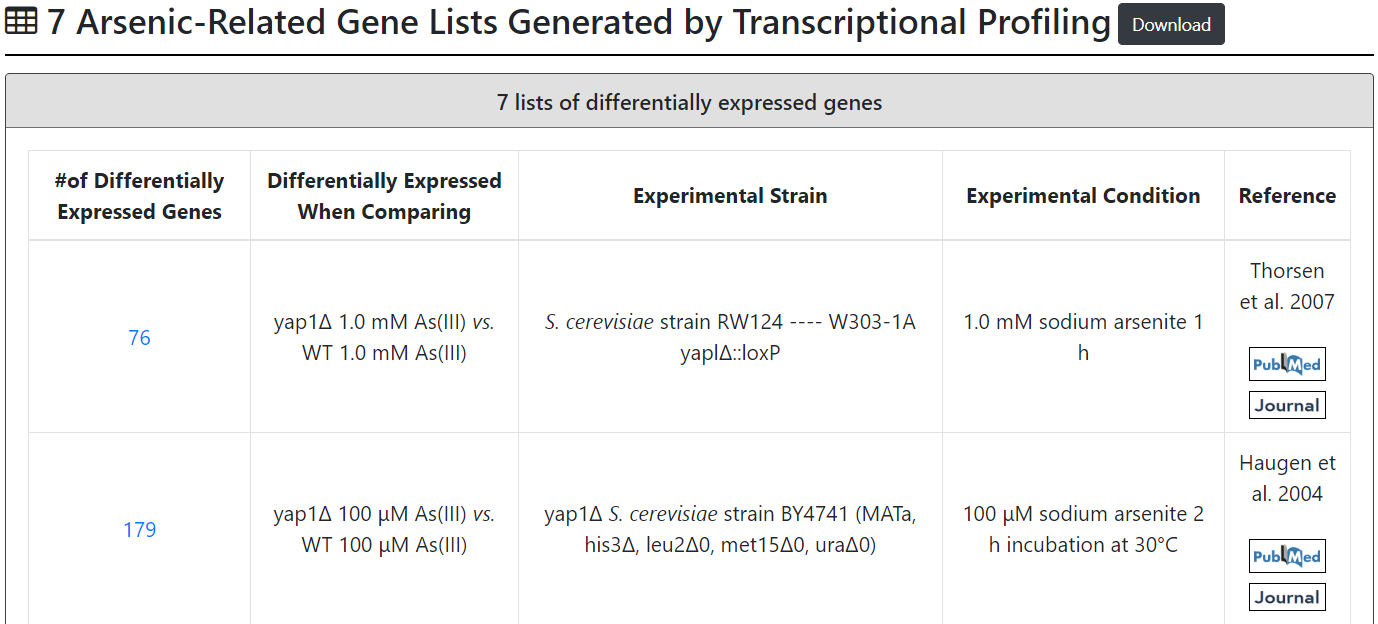
| Thorsen et al. 2007 | 76 | yap1Δ 1.0 mM As(III) vs. WT 1.0 mM As(III) | S. cerevisiae RW124-W303-1A yaplΔ::loxP | 1.0 mM sodium arsenite for 1 h |
| Haugen et al. 2004 | 179 | yap1Δ 100 μM As(III) vs. WT 100 μM As(III) | yap1Δ of S. cerevisiae BY4741 (MATa, his3Δ, leu2Δ0, met15Δ0, uraΔ0) | 100 μM sodium arsenite for 2 h |
| Haugen et al. 2004 | 415 | rpn4Δ 100 μM As(III) vs. WT 100 μM As(III) | rpn4Δ S. cerevisiae BY4741 (MATa, his3Δ, leu2Δ0, met15Δ0, uraΔ0) | 100 μM sodium arsenite for 2 h |
| Haugen et al. 2004 | 875 | arr1Δ 100 μM As(III) vs. WT 100 μM As(III) | arr1Δ of S. cerevisiae BY4741 (MATa, his3Δ, leu2Δ0, met15Δ0, uraΔ0) | 100 μM sodium arsenite for 2 h |
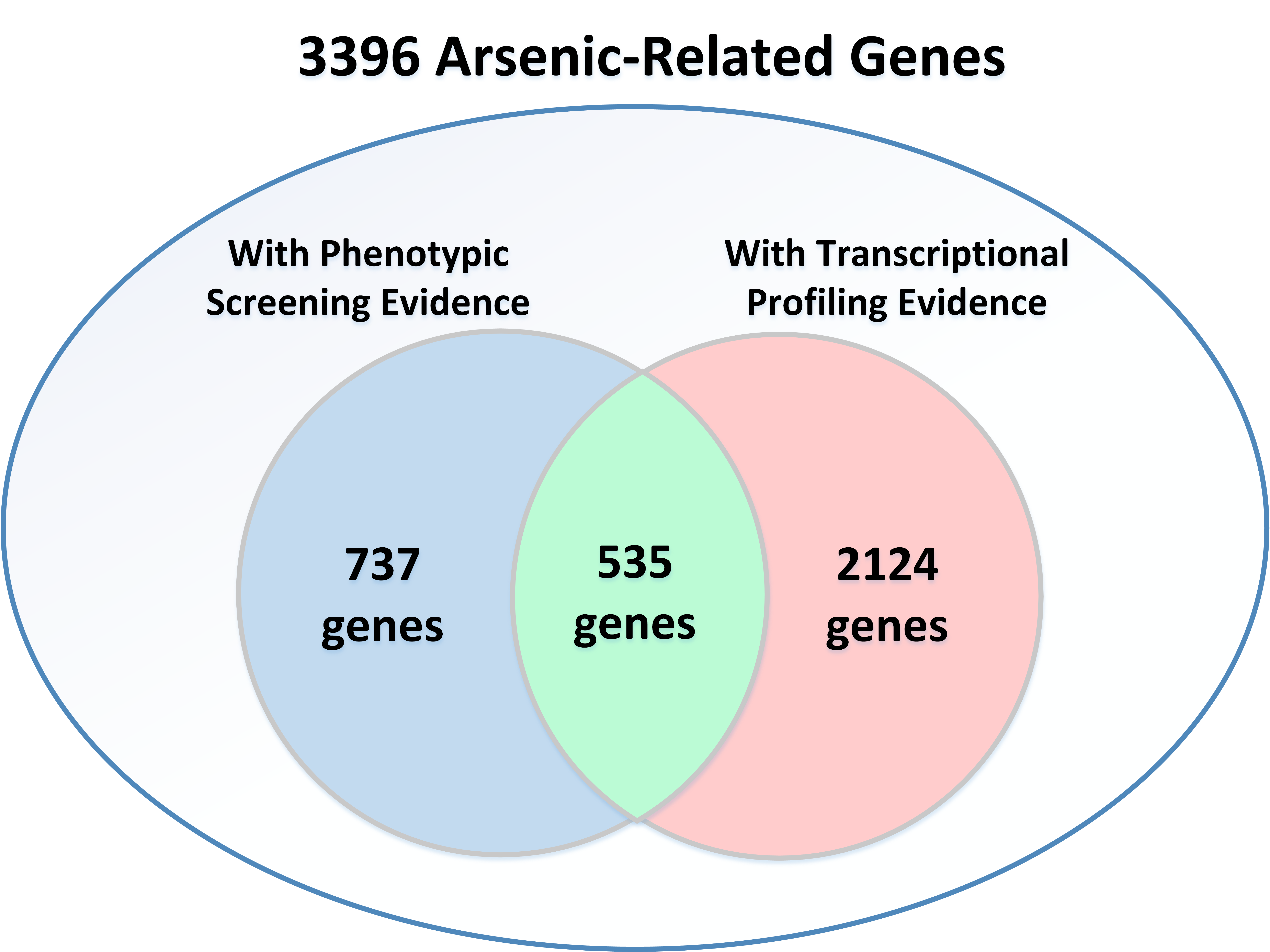
Among the 3396 arsenic-related genes, 535 are supported by both PS and TP, 737 are supported only by PS, and 2124 are supported only by TP.
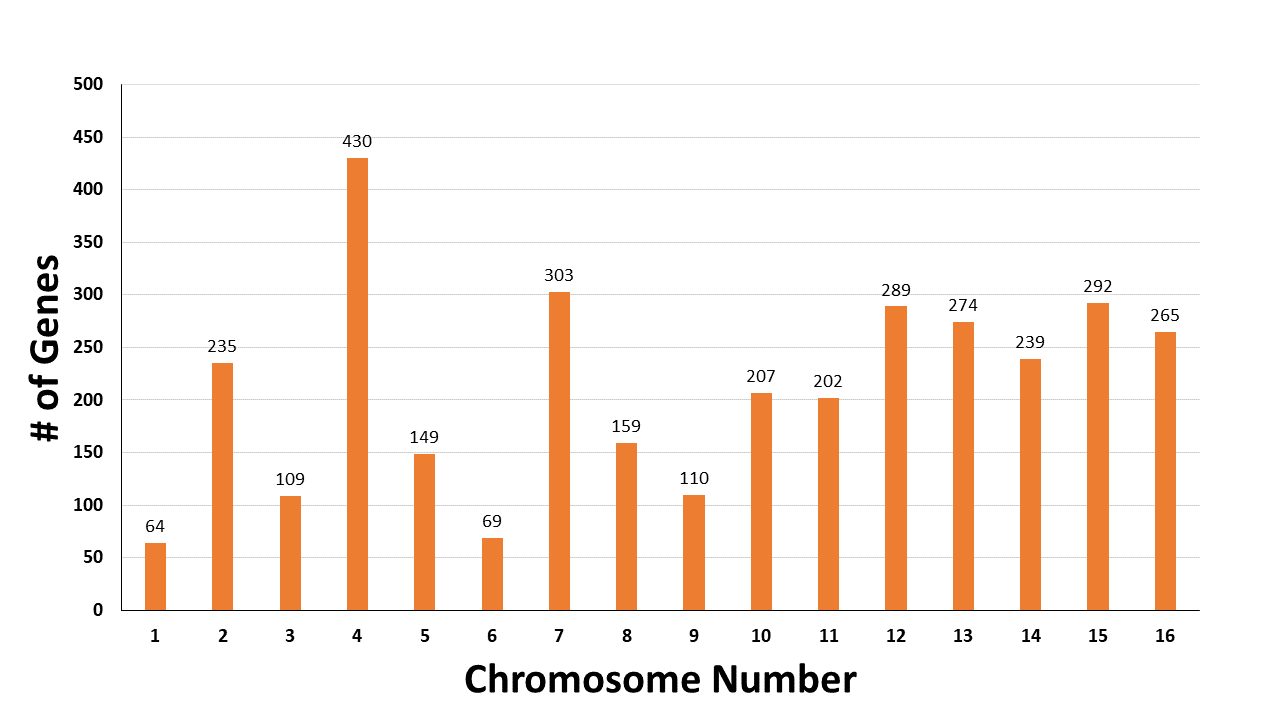
The distribution of these 3396 arsenic-related genes on different chromosomes is shown.
Testing the enrichment of arsenic-related genes in the input genes
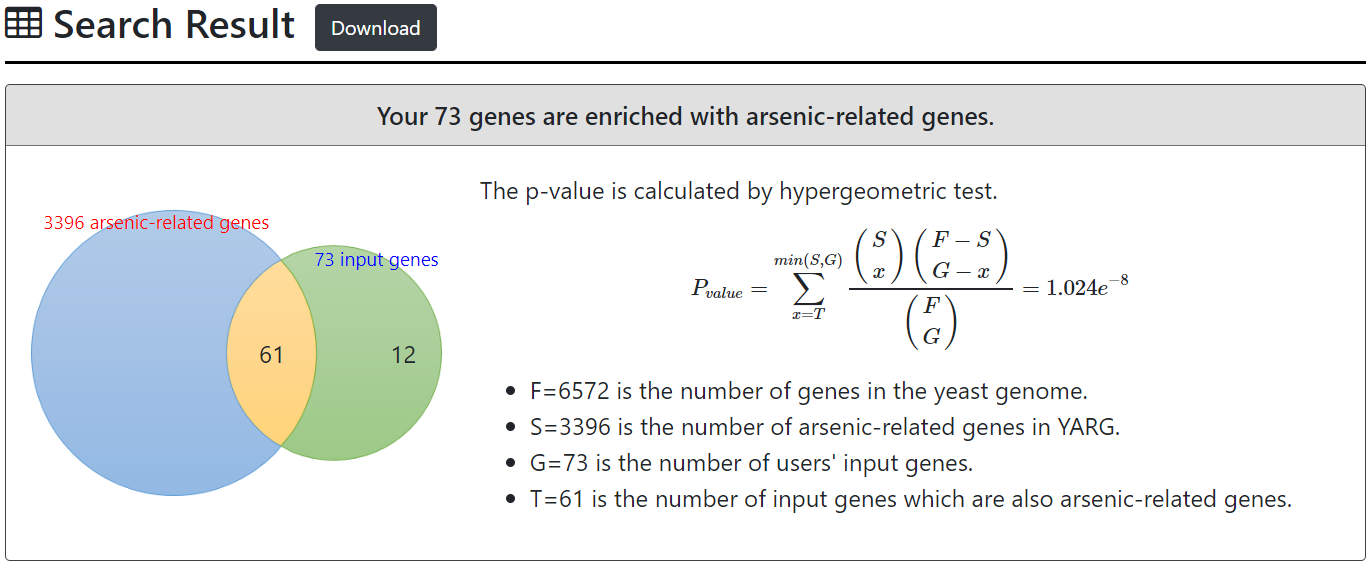
For users' input genes, YARG tests whether they are enriched with arsenic-related genes. The p-value is calculated using hypergeometric test as follows.
$$P_{value} = \sum^{min(S,G)}_{x=T} \frac{\left(\begin{matrix}S\\x\end{matrix} \right)\left(\begin{matrix}F-S\\G-x\end{matrix}\right)}{\left(\begin{matrix}F\\G\end{matrix}\right)}$$
where F=6572 is the number of genes in the yeast genome, S=3396 is the number of arsenic-related genes in YARG, G is the number of users’ input genes, and T is the number of input genes which are also arsenic-related genes.
Database interface
YARG provides two search modes
First search mode:
Users can input a gene name.
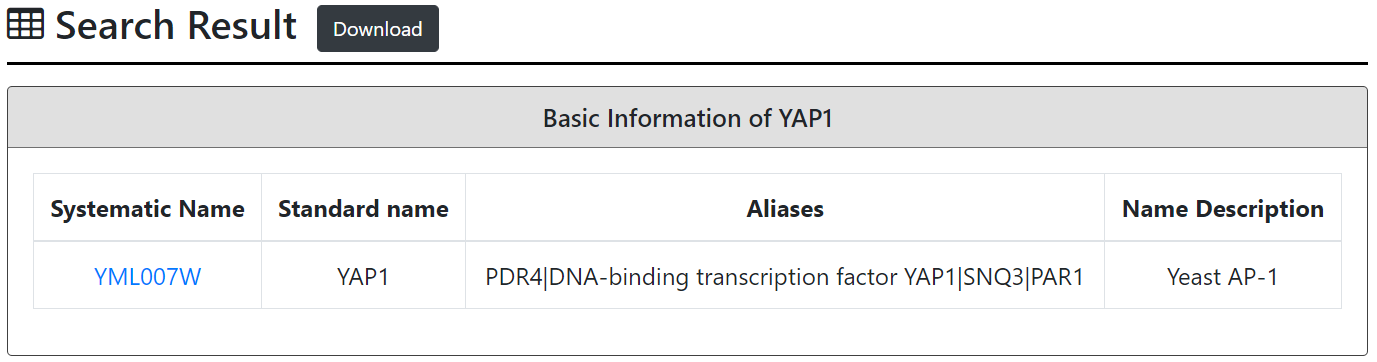
After submission, YARG returns a page showing the basic information of the input gene and links to YeastMine to see the homology information such as human homologs, fungal homologs, non-fungal homologs, functional complementation and paralogs.

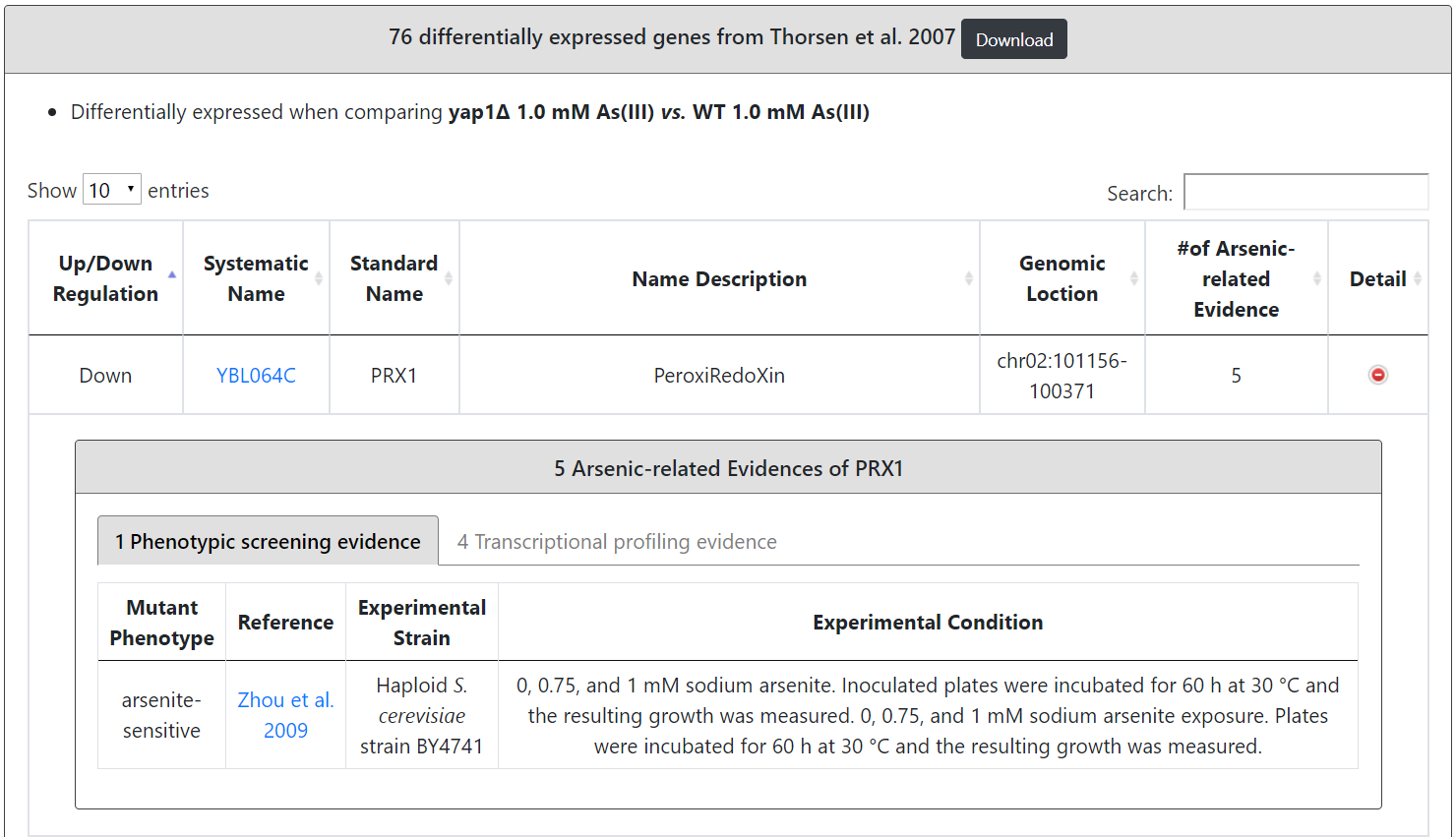
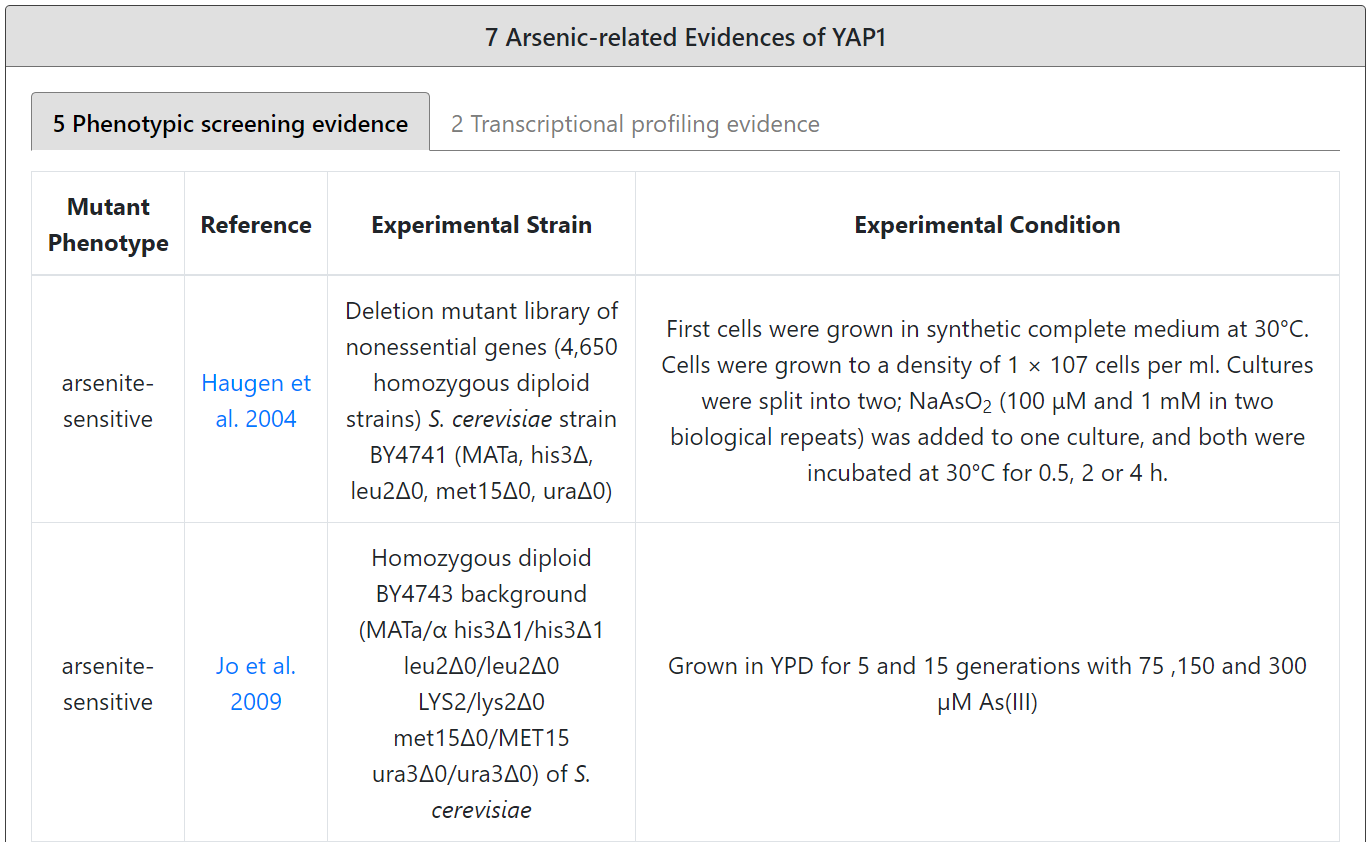
If the input gene is an arsenic-related gene, the details (experimental strain, experimental condition and reference) of the experimental evidence (phenotypic screening and/or transcriptional profiling) are provided.
Second search mode:
Users can input a list of genes.
After submission, YARG uses the hypergeometric test to test whether the input genes are enriched with arsenic-related genes.
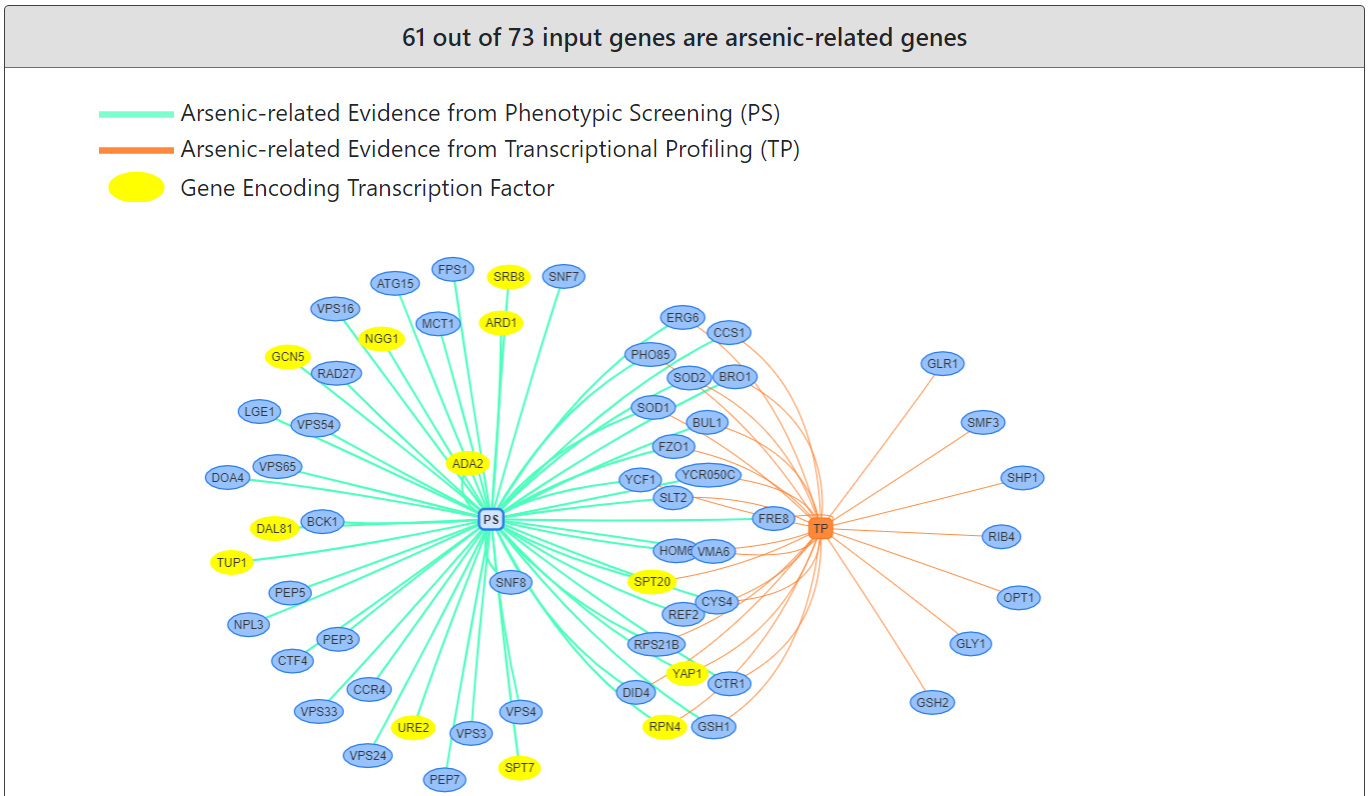
YARG also provides a figure and a table to show which input genes are arsenic-related genes and the number of supporting evidence.
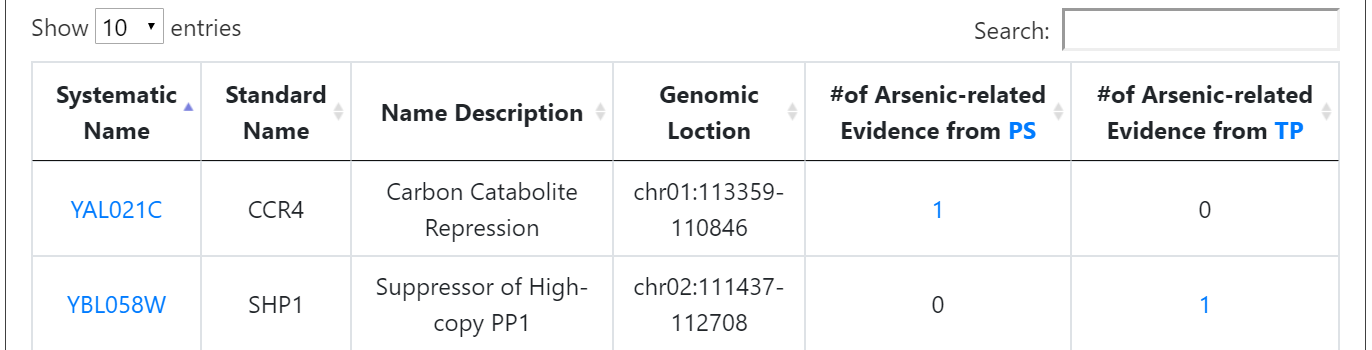
The details (experimental strain, experimental condition and reference) of the supporting evidence are also shown.
YARG provides three browse modes
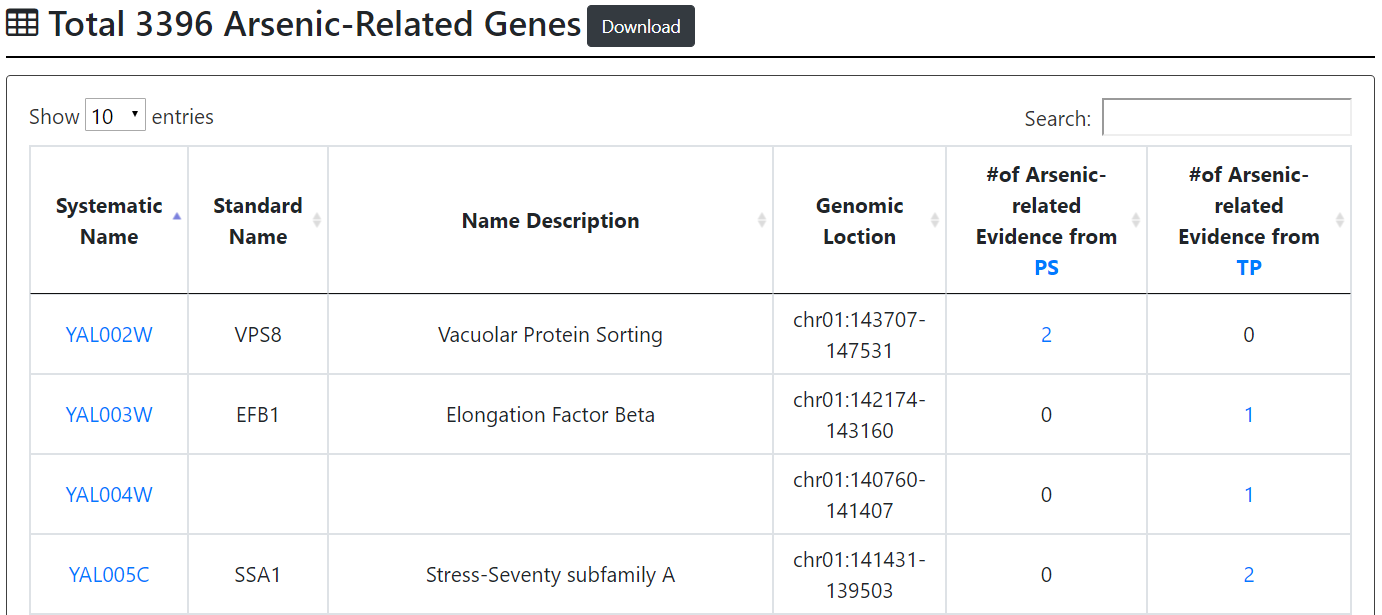
First browse mode:
Users can browse 3396 arsenic-related genes. For each gene, YARG provides the systematic name, standard name, name description, genomic location, the number of arsenic-related evidence from PS, and the number of arsenic-related evidence from TP.
Second browse mode:
Users can browse 13 arsenic-related gene lists generated by phenotypic screening. These 13 arsenic-related gene lists consist of 1 mutant gene list of arsenate-sensitive phenotypes, 3 mutant gene lists of arsenite-resistant phenotypes and 9 mutant gene lists of arsenite-sensitive phenotypes.

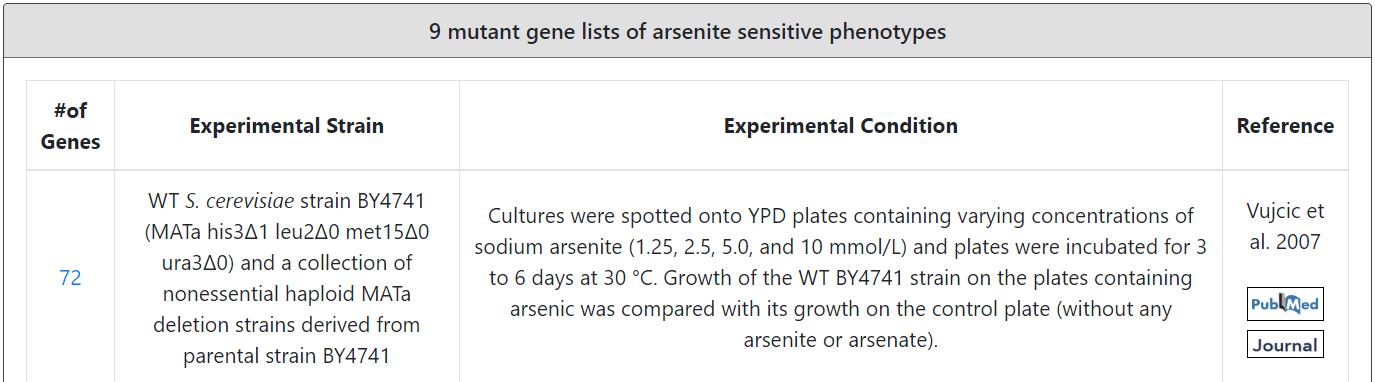
Third browse mode:
Users can browse 7 arsenic-related gene lists generated by transcriptional profiling. These 7 arsenic-related gene lists consist of (i) 3 lists of genes which are differentially expressed between WT and WT under arsenic exposure and (ii) 4 lists of genes which are differentially expressed between WT and the transcription factor mutant (arr1Δ, rpn4Δ or yap1Δ) both under arsenic exposure.